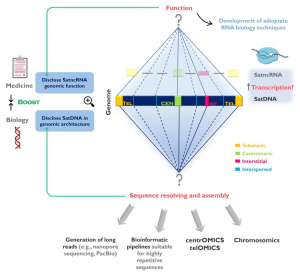
Sandra Louzada, Mariana Lopes, Daniela Ferreira, Filomena Adega, Ana Escudeiro, Margarida Gama-Carvalho, and Raquel Chaves
Doi: 10.3390/genes11010072
Cited as: Louzada S, Lopes M, Ferreira D, Adega F, Escudeiro A, Gama-Carvalho M, Chaves R (2020) Decoding the Role of Satellite DNA in Genome Architecture and Plasticity – An Evolutionary and Clinical Affair. Genes 11(1), 72; https://doi.org/10.3390/genes11010072.