Elisabete Santos, Maria Manuela Matos, and Cesar Benito
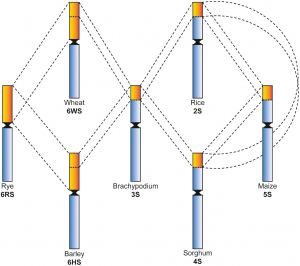
Aluminium (Al) toxicity is the major constraint for crop productivity in acid soils. Wild rye species (Secale spp.) exhibit high Al tolerance, being a good source of genes related to this trait. The Alt1 locus located on the 6RS chromosome arm is one of the four main loci controlling Al tolerance in rye and is known to harbour major genes but, so far, none have been found. Through synteny among the short arm of the rye chromosome 6R and the main grass species, we found a candidate MATE gene for the Atl1 locus, later named ScMATE3, which was isolated and characterized in different Secale species. The sequence comparisons revealed both intraspecific and interspecific variability, with high sequence conservation in the Secale genus. SNP with replacement substitution that changed the structure of the protein and can be involved in the Al tolerance trait were found in ScMATE3 gene. The predicted subcellular localization of ScMATE3 is the vacuolar membrane which, together with the phylogenetic relationships performed with other MATE genes of the Poaceae related to Al detoxification, suggest involvement of ScMATE3 in an internal tolerance mechanism. Moreover, expression studies of this gene in rye corroborate its contribution in some Al resistance mechanisms. The ScMATE3 gene is located on the 6RS chromosome arm between the same markers in which the Alt1 locus is involved in Al resistance mechanisms in rye, thus being a good candidate gene for this function.
Doi: 10.1111/plb.13107
Cited as: Santos E, Matos M, Benito C (2020). Isolation and characterization of a new MATE gene located in the same chromosome arm of the aluminium tolerance (Alt1) rye locus. Plant Biology 22, 691–700; https://doi.org/10.1111/plb.13107.